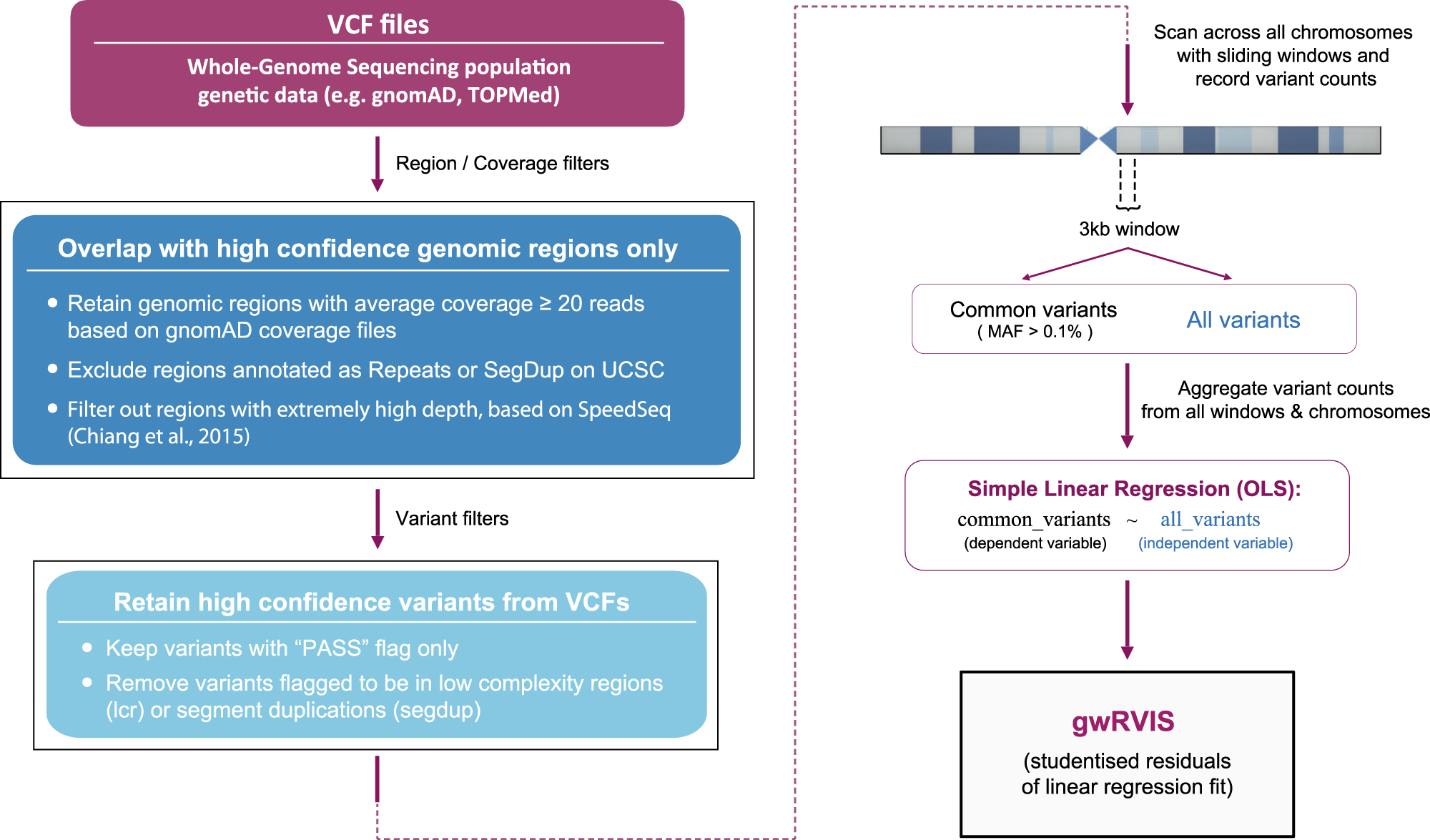
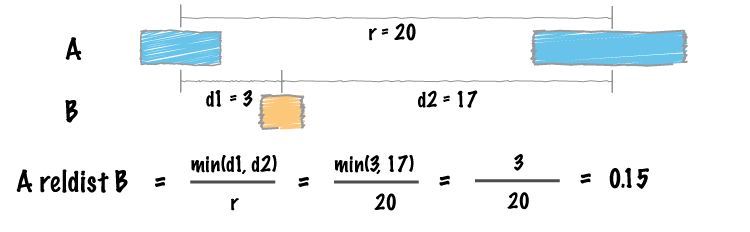
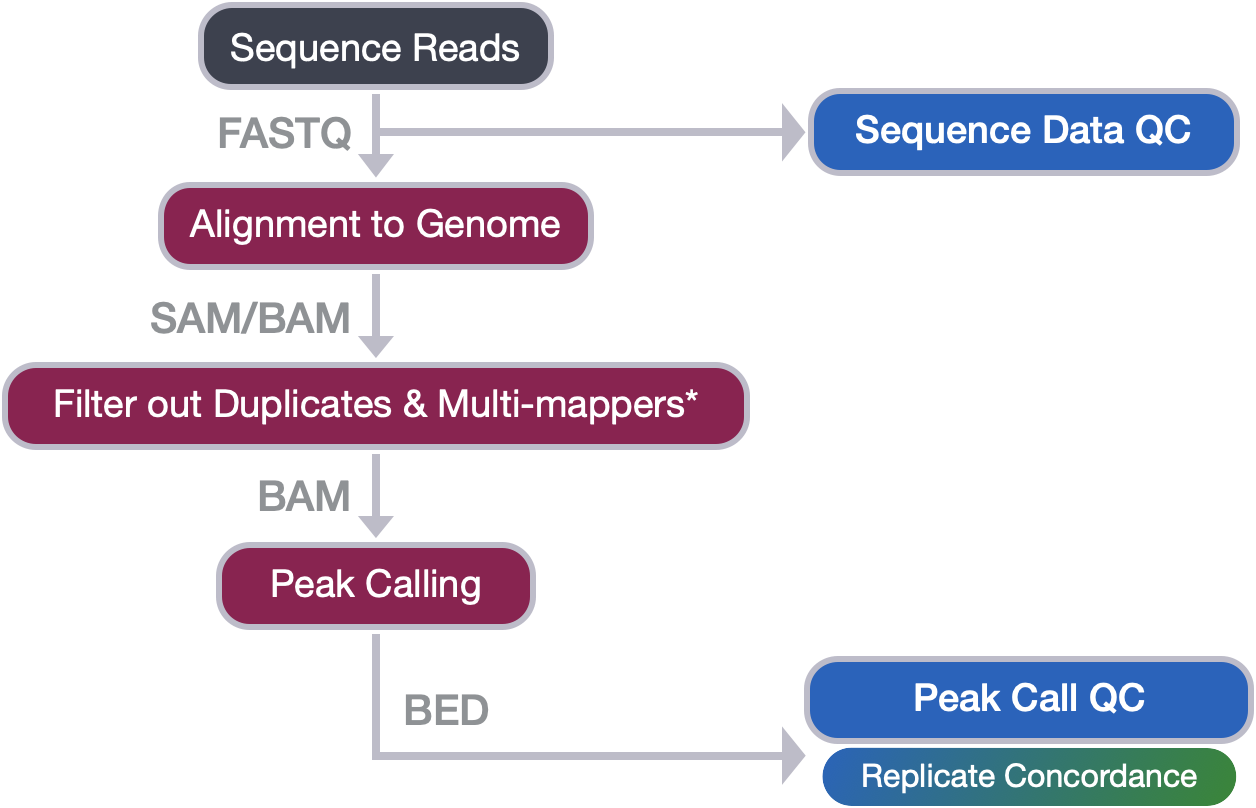
Protocol for unbiased, consolidated variant calling from whole exome sequencing data - ScienceDirect

Multiplex-GAM: genome-wide identification of chromatin contacts yields insights overlooked by Hi-C | Nature Methods

Highly sensitive single-cell chromatin accessibility assay and transcriptome coassay with METATAC | PNAS
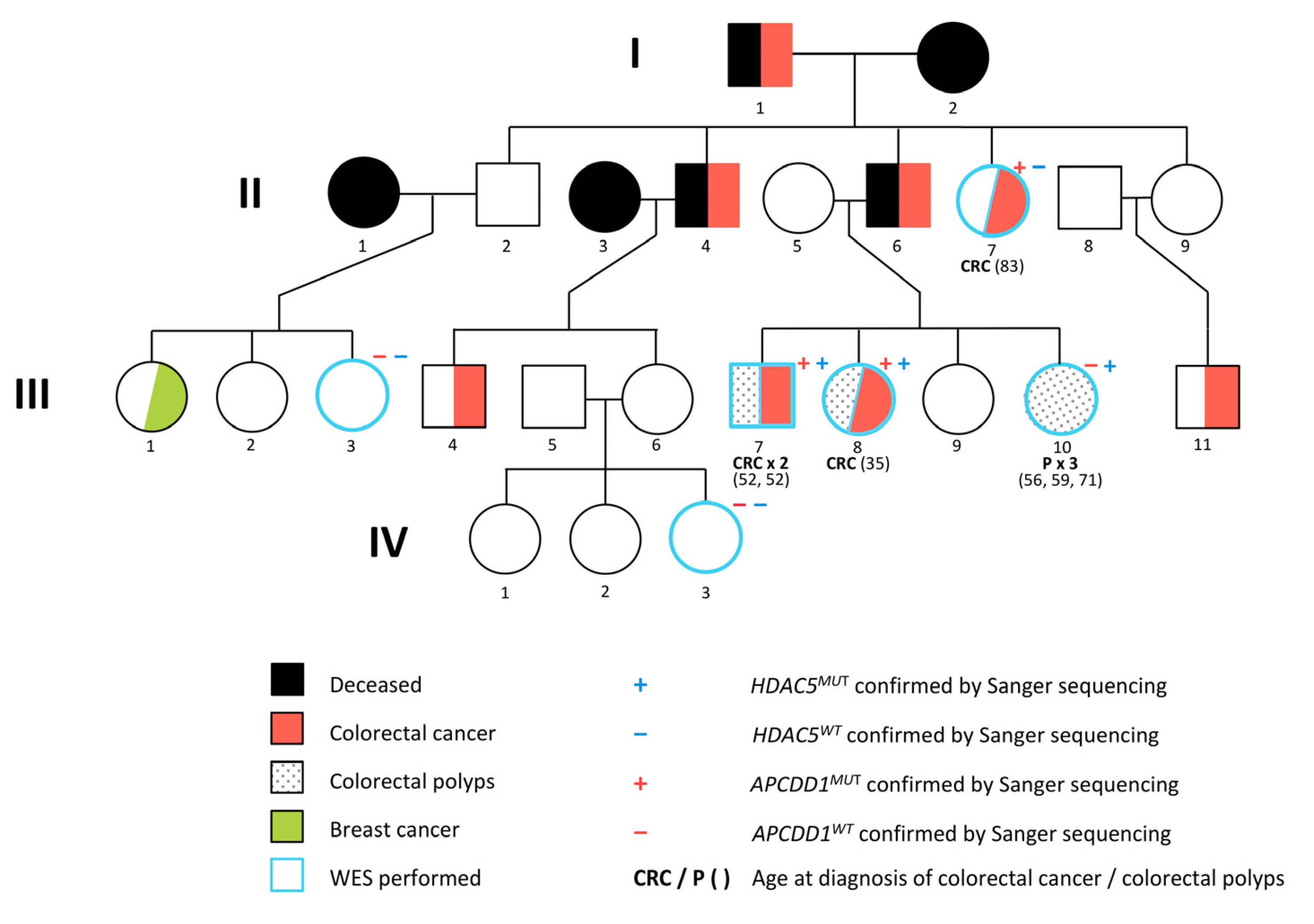
IJMS | Free Full-Text | Whole Exome Sequencing Identifies APCDD1 and HDAC5 Genes as Potentially Cancer Predisposing in Familial Colorectal Cancer

Genomic evidence for homoploid hybrid speciation in a marine mammal apex predator | Science Advances

Butterfly eyespots evolved via cooption of an ancestral gene-regulatory network that also patterns antennae, legs, and wings | PNAS